Research Projects
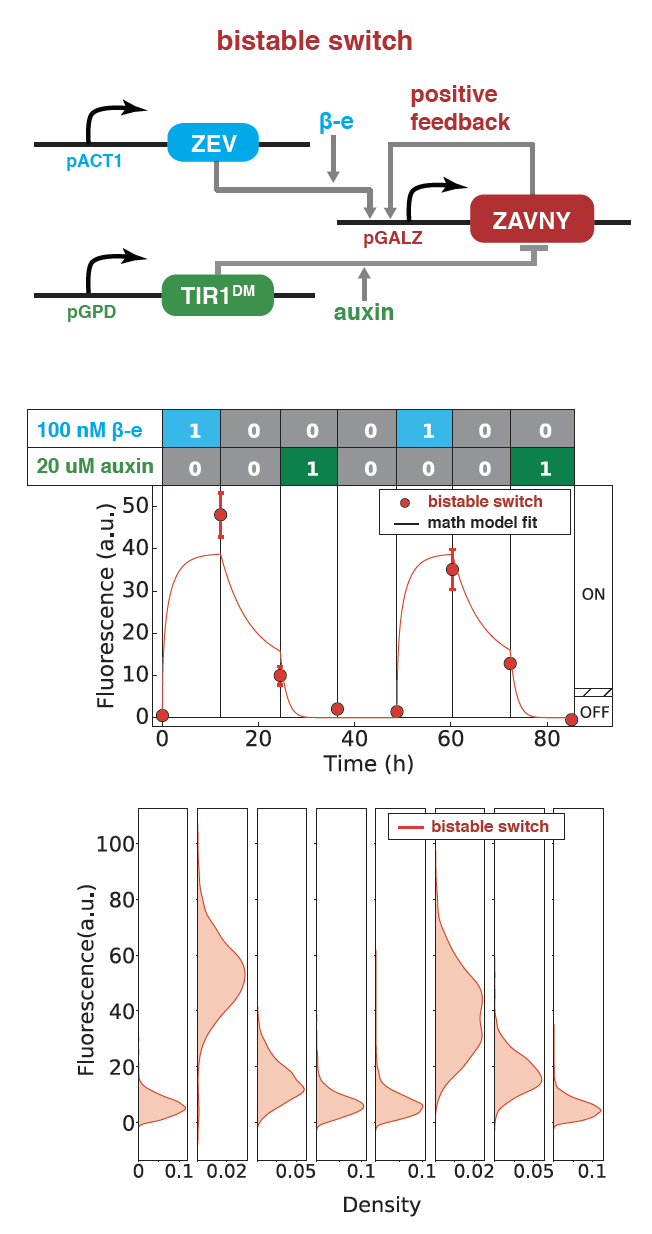
Synthetic Bistability and Differentiation in Yeast
Engineered systems that control cellular differentiation and pattern formation are essential for applications like tissue engineering, biomaterial fabrication, and synthetic ecosystems. Synthetic circuits that can take on multiple states have been made to engineer multicellular systems. However, how to use these states to drive interesting cellular behavior remains challenging. I built a cellular differentiation program involving a novel synthetic bistable switch coupled to an antibiotic resistance gene that affects growth in yeast (S. cerevisiae). The switch is composed of a positive feedback loop involving a novel transcription factor and can be switched ON and OFF via two different transient inducer inputs. By further coupling the bistable switch with an antibiotic resistance gene, we obtained a growth differentiation circuit, where yeast cells can be switched to stable HIGH or LOW growth rate states via transient inducer inputs. This work demonstrates a rationally designed and experimentally validated cellular differentiation behavior in yeast.
Related work
- Y. Yang, J. L. Nemhauser, E. Klavins. Synthetic Bistability and Differentiation in Yeast. ACS synthetic biology, 2019.
- Y. Yang, E. Klavins. Synthetic bistability and antibiotic resistance memory in S. cerevisiae. Talk, Molecular Programming Project Workshop, Boston, USA, 2016.
- Y. Yang, E. Klavins. Design and construction of a bistable switch in yeast. Poster and Lightning Talk, Molecular Programming Project Workshop, San Francisco, USA, 2015.
- Y. Yang, E. Klavins. Design and construction of a switchable bistable switch in yeast. Poster, SEED 2015, Boston, USA, 2015.
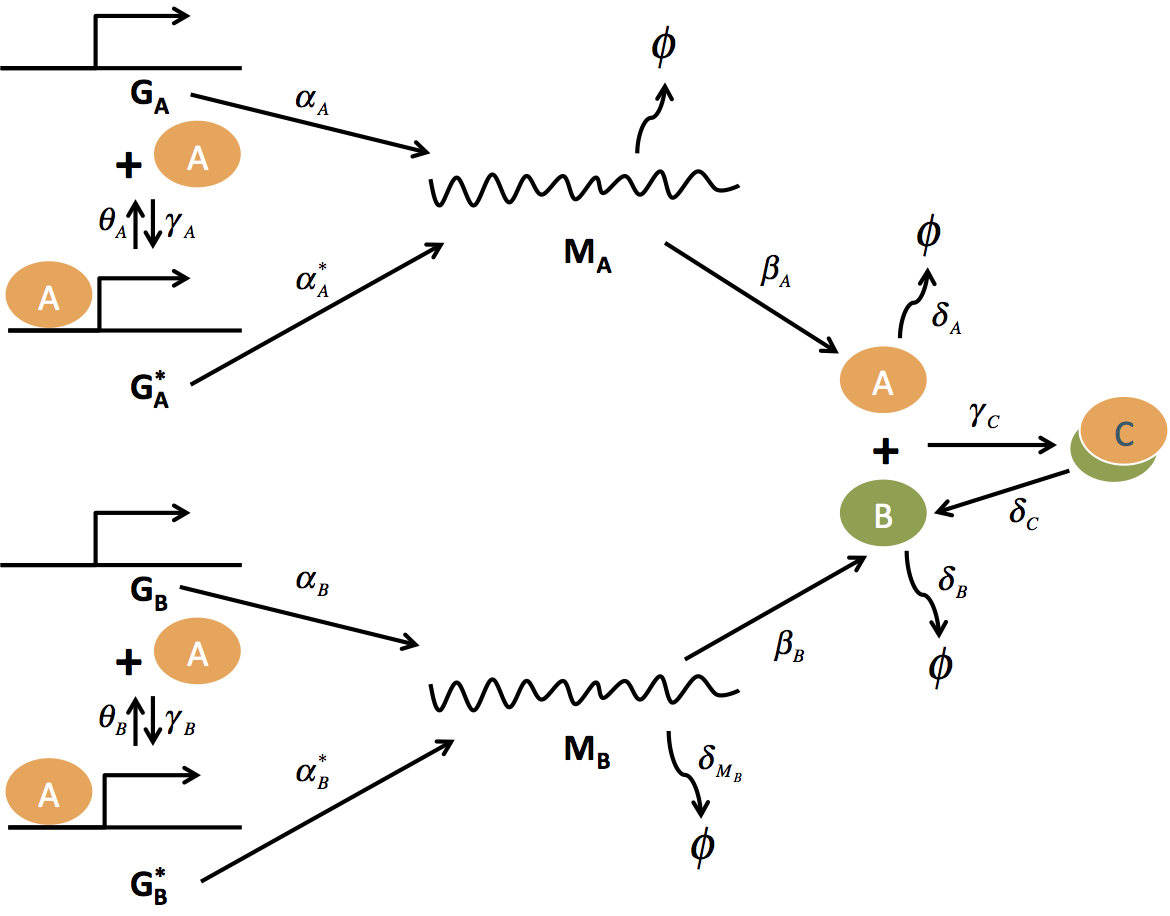
A plant hormone induced oscillator in yeast
Biochemical oscillators are fundamental to understanding cell physiology, and present an ongoing challenge in building synthetic living systems. In synthetic biology, researchers have built a number of oscillators over the last decade resulting in various designs in organisms from E. coli to mammalian cells. However, especially in eukaryotic cells, a robust, tunable, and modular oscillator technology has yet to be demonstrated. In this project, I designed and analyzed a plant hormone induced oscillator in yeast. I developed mathematical model and showed that the oscillator could be achieved by tuning two key parameters. Currently I am working on experimental realizing this oscillator.
Related work
- Y. Yang, E. Klavins. Design and analysis of a plant hormone induced oscillator in yeast. Abstract and talk, CSHA Synthetic Biology, Suzhou, China, 2014.
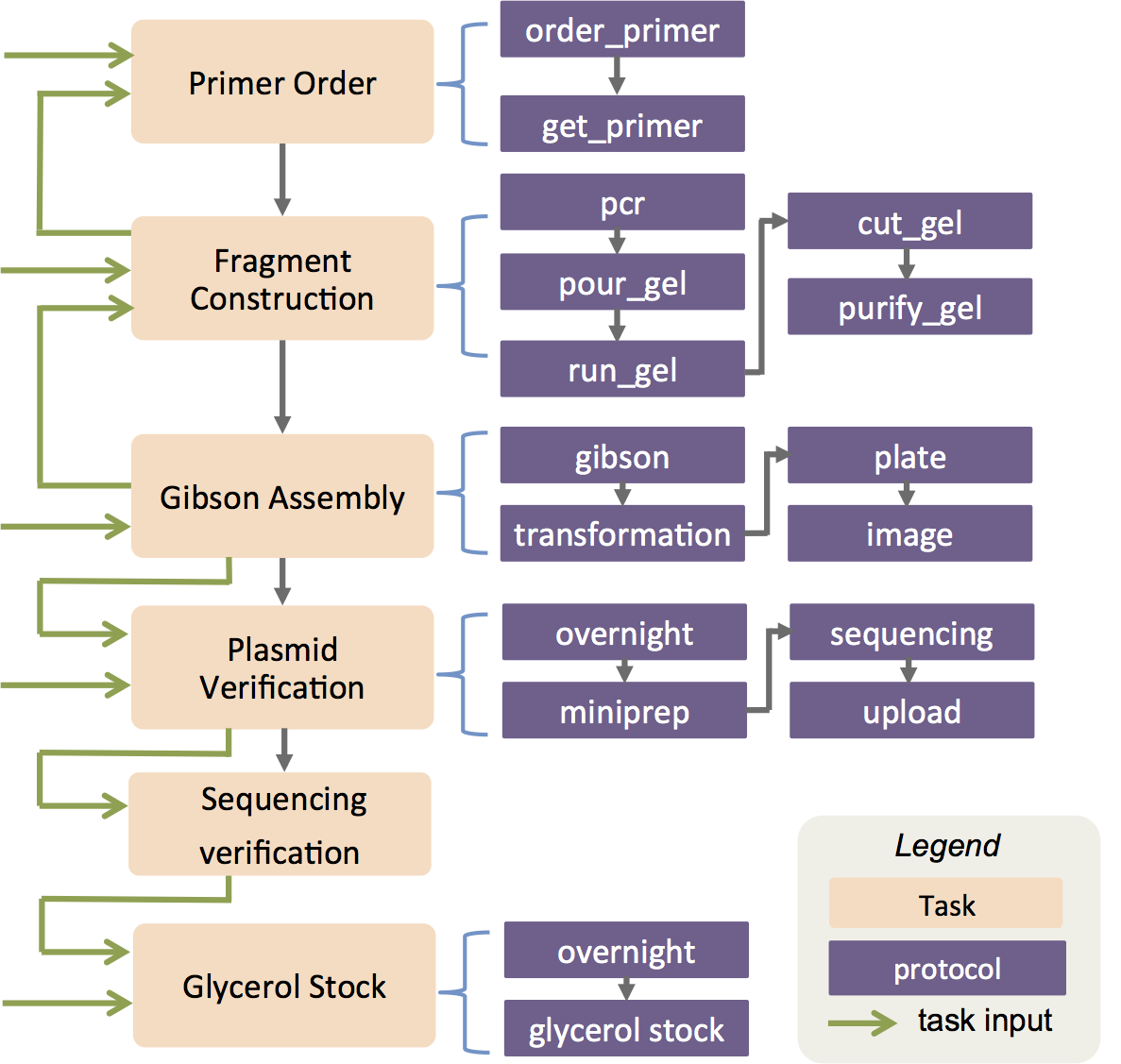
The Aquarium cloning workflows
Aquarium is a smart laboratory operating system invented in the Klavins Lab where researchers can specify how to manipulate items that stored in the inventory using protocols and workflows written in computer codes. Adapted from the standard molecular cloning workflows, I wrote a majority of the codes in Ruby implementing two core molecular cloning workflows: plasmid construction and yeast cloning. These workflows take and batch users' inputs to the workflow and semi-automatically schedule the entire cloning process from ordering primers to Sanger sequencing verifications, resulting a nearly effortless cloning process in Aquarium. This technology also led to the startup Aquarium where I worked as an engineer to build out the 21st century laboratory operating system, enabling reproducible and transferable workflows in biology labs and more.
Related work
- J. Vrana, O. de Lange, Y. Yang, G. Newman, A. Saleem, A. Miller, C. Cordray, S. Halabiya, M. Parks, E. Lopez, S. Goldberg, B. Keller, D. Strickland, E. Klavins. Aquarium: open-source laboratory software for design, execution and data management, Synthetic Biology, 2021.
- Aquarium Lab in the Cloud. Engineering research projects haul in CoMotion Innovation Fund dollars. News, University of Washington, 2015.
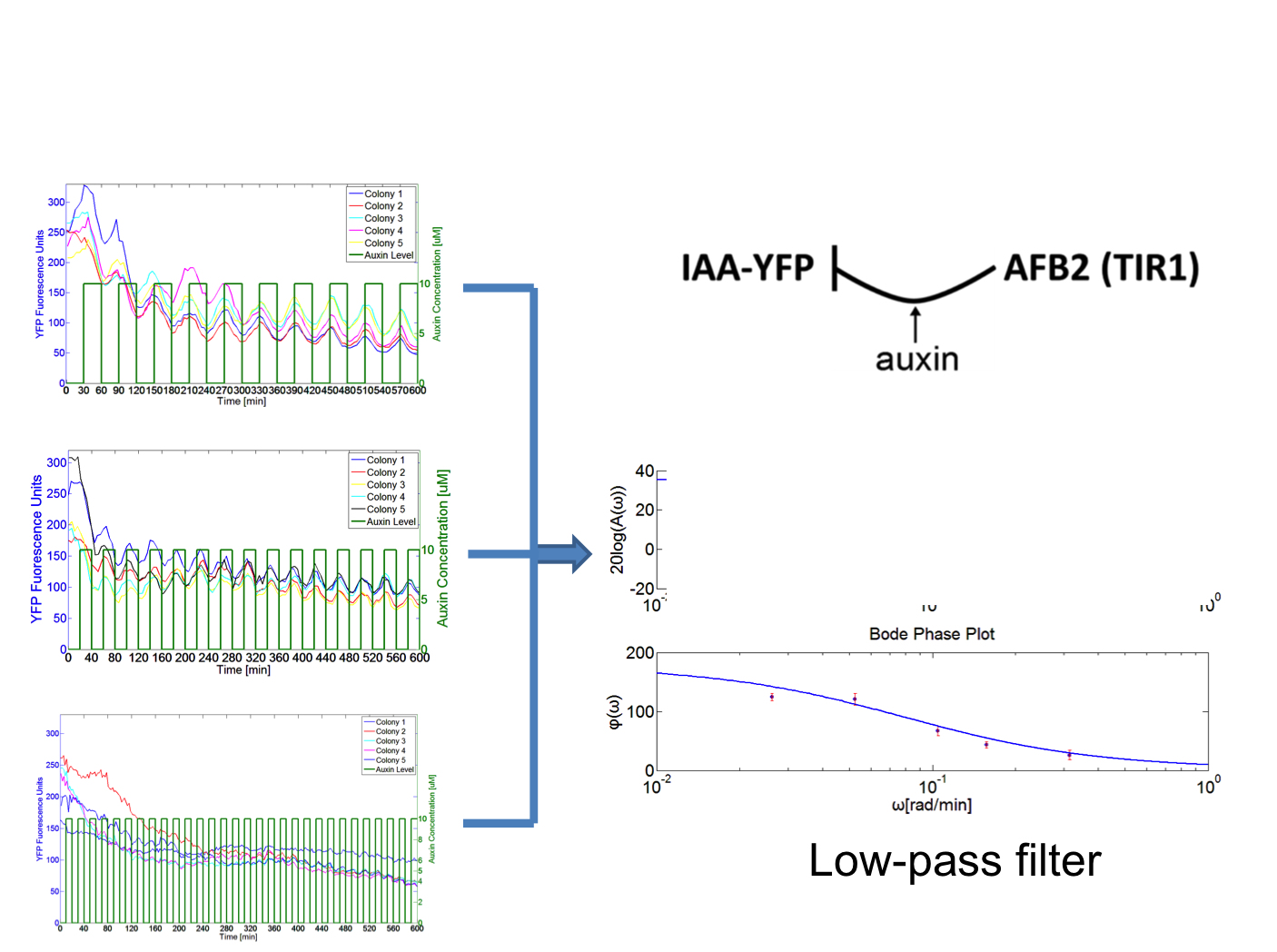
Frequency response for a synthetic auxin signaling pathway in yeast
I developed an experimental process in the Klavins Lab to interrogate frequency response for a synthetic auxin signaling pathway in yeast. The approach was using a microfluidic device called CellASIC to grow the yeast cell, where one can control the system precisely with different frequencies of auxin inputs and observe using fluorescence microscopy over time. I wrote Matlab based software to do image processing on the time course fluorescence images, which resulted in quantified time-course fluorescence data on the cells. Using the data, I generated a Bode plot describing the frequency response of signaling pathway. The result showed that this pathway works as a low-pass filter.
Related work
- Y. Yang, E. Klavins. Frequency domain signal processing in biological system. Poster and Lightning Talk, Molecular Programming Project Workshop, Oxnard, CA, USA, 2013.